Exercise 4: Metropolis-Hastings algorithm(s) for the historical application (Beta-Bernoulli model)
Using the historical example, program an independent Metropolis-Hastings algorithm to estimate the posterior distribution of parameter
Program a function that computes the numerator of the posterior density, which can be written
post_num_hist <- function(theta, log = FALSE) { n <- 493472 # the data S <- 241945 # the data if (log) { num <- S * log(theta) + (n - S) * log(1 - theta) # the **log** numerator of the posterior } else { num <- theta^S * (1 - theta)^(n - S) # the numerator of the posterior } return(num) # the output of the function } post_num_hist(0.2, log = TRUE)## [1] -445522.1post_num_hist(0.6, log = TRUE)## [1] -354063.6post_num_hist(0.2)## [1] 0post_num_hist(0.6)## [1] 0Program the corresponding Metropolis-Hastings algorithm, returning a vector of size
myMH <- function(niter, post_num) { x_save <- numeric(length = niter) #create a vector of 0s # of length niter to store the sampled values alpha <- numeric(length = niter) #create a vector of 0s # of length niter to store the acceptance probabilities # initialise x0 x <- runif(n = 1, min = 0, max = 1) # acceptance-rejection loop for (t in 1:niter) { # sample y from the proposal (here uniform prior) y <- runif(n = 1, min = 0, max = 1) # compute the acceptance-rejection probability alpha[t] <- min(1, exp(post_num(y, log = TRUE) - post_num(x, log = TRUE))) # accept or reject u <- runif(1) if (u <= alpha[t]) { x_save[t] <- y } else { x_save[t] <- x } # update the current value x <- x_save[t] } return(list(theta = x_save, alpha = alpha)) }Compare the posterior density obtained with this Metropolis-Hastings algorithm over 2000 iterations to the theoretical one (the theoretical density can be obtained with the
Rfunctiondbeta(x, 241945 + 1, 251527 + 1)and represented with theRfunctioncurve(..., from = 0, to = 1, n = 10000)). Mindfully discard the first 500 iterations of your Metropolis-Hastings algorithm in order to reach the Markov chain convergence before constructing your Monte Carlo sample. Comment those results, especially in light of the acceptance probabilities computed throughout the algorithm, as well as the different sampled values forsampleMH <- myMH(2000, post_num = post_num_hist) par(mfrow = c(2, 2)) plot(density(sampleMH$theta[-c(1:500)]), col = "red", xlim = c(0, 1), ylab = "Posterior probability density", xlab = expression(theta), main = "") curve(dbeta(x, 241945 + 1, 251527 + 1), from = 0, to = 1, n = 10000, add = TRUE) legend("topright", c("M-H", "theory"), col = c("red", "black"), lty = 1) plot(density(sampleMH$theta[-c(1:500)]), col = "red", ylab = "Posterior probability density", xlab = expression(theta), main = "Zoom") curve(dbeta(x, 241945 + 1, 251527 + 1), from = 0, to = 1, n = 10000, add = TRUE) legend("topright", c("M-H", "theory"), col = c("red", "black"), lty = 1) plot(sampleMH$alpha, type = "h", xlab = "Iteration", ylab = "Acceptance Probability", ylim = c(0, 1), col = "springgreen") plot(sampleMH$theta, type = "l", xlab = "Iteration", ylab = expression(paste("Sampled value for ", theta)), ylim = c(0, 1))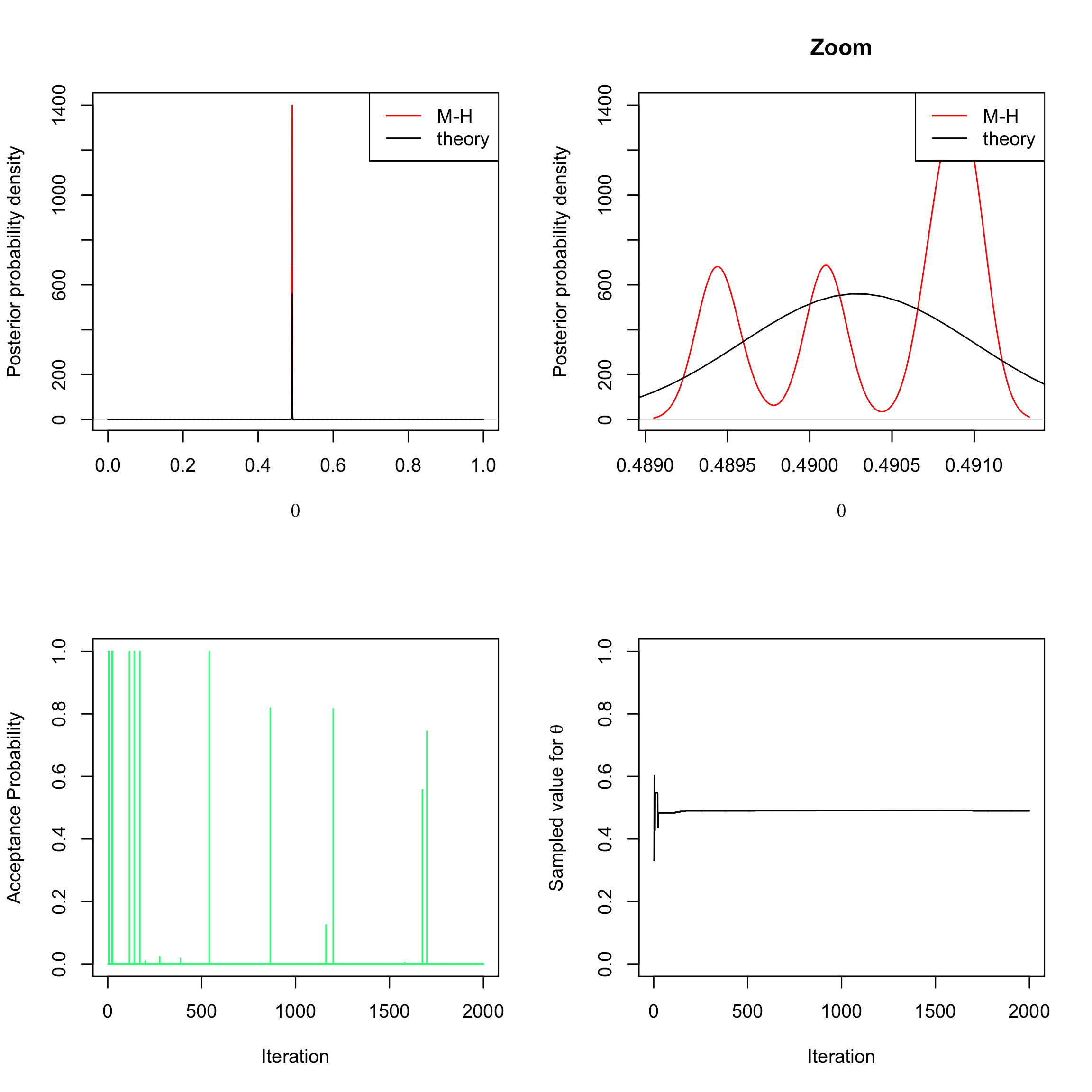
Now imagine we only observe
post_num_beta <- function(theta, a = 3, b = 3, log = TRUE) { n <- 100 #number of trials (births) S <- 49 #number of success (feminine births) if (log) { num <- (a + S - 1) * (log(theta)) + (b + n - S - 1) * log(1 - theta) } else { num <- theta^(a + S - 1) * (1 - theta)^(b + n - S - 1) } return(num) } myMH_betaprior <- function(niter, post_num, a = 3, b = 3) { x_save <- numeric(length = niter) # create a vector of 0s of length niter to store the sampled values alpha <- numeric(length = niter) # create a vector of 0s of length niter to store the acceptance probabilities # initialise x x <- runif(n = 1, min = 0, max = 1) # acceptance-rejection loop for (t in 1:niter) { # sample a value from the proposal (beta prior) y <- rbeta(n = 1, a, b) # compute acceptance-rejection probability alpha[t] <- min(1, exp(post_num(y, a = a, b = b, log = TRUE) - post_num(x, a = a, b = b, log = TRUE) + dbeta(x, a, b, log = TRUE) - dbeta(y, a, b, log = TRUE))) # acceptance-rejection step u <- runif(1) if (u <= alpha[t]) { x <- y # acceptance of y as new current value } # saving the current value of x x_save[t] <- x } return(list(theta = x_save, alpha = alpha)) } sampleMH <- myMH_betaprior(10000, post_num = post_num_beta) par(mfrow = c(2, 2)) plot(density(sampleMH$theta[-c(1:500)]), col = "red", xlim = c(0, 1), ylab = "Posterior probability density", xlab = expression(theta), main = "") curve(dbeta(x, 49 + 1, 51 + 1), from = 0, to = 1, add = TRUE) legend("topright", c("M-H", "theory"), col = c("red", "black"), lty = 1) plot.new() plot(sampleMH$alpha, type = "h", xlab = "Iteration", ylab = "Acceptance probability", ylim = c(0, 1), col = "springgreen") plot(sampleMH$theta, type = "l", xlab = "Iteration", ylab = expression(paste("Sampled value for ", theta)), ylim = c(0, 1))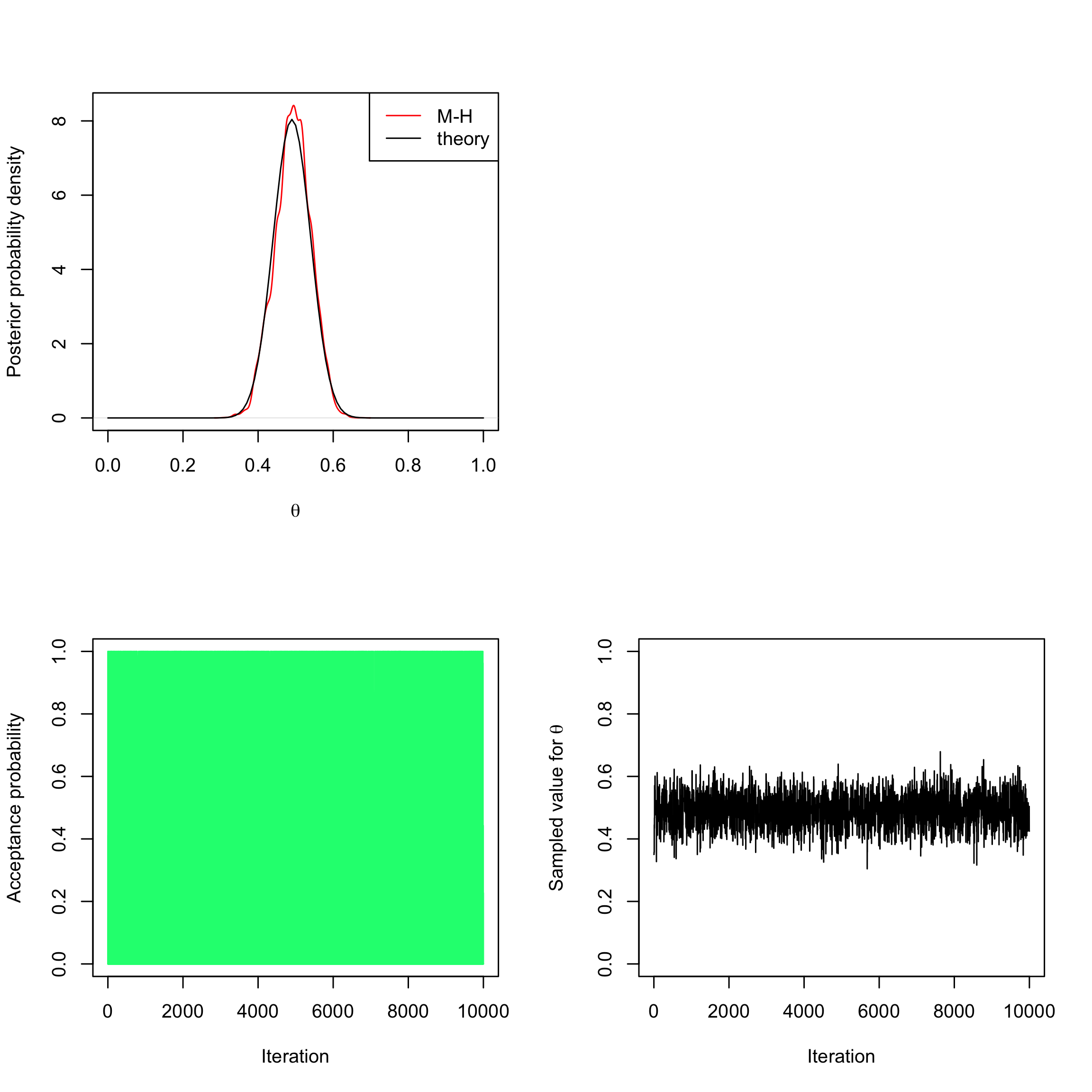
Using the data from the historical example and with a
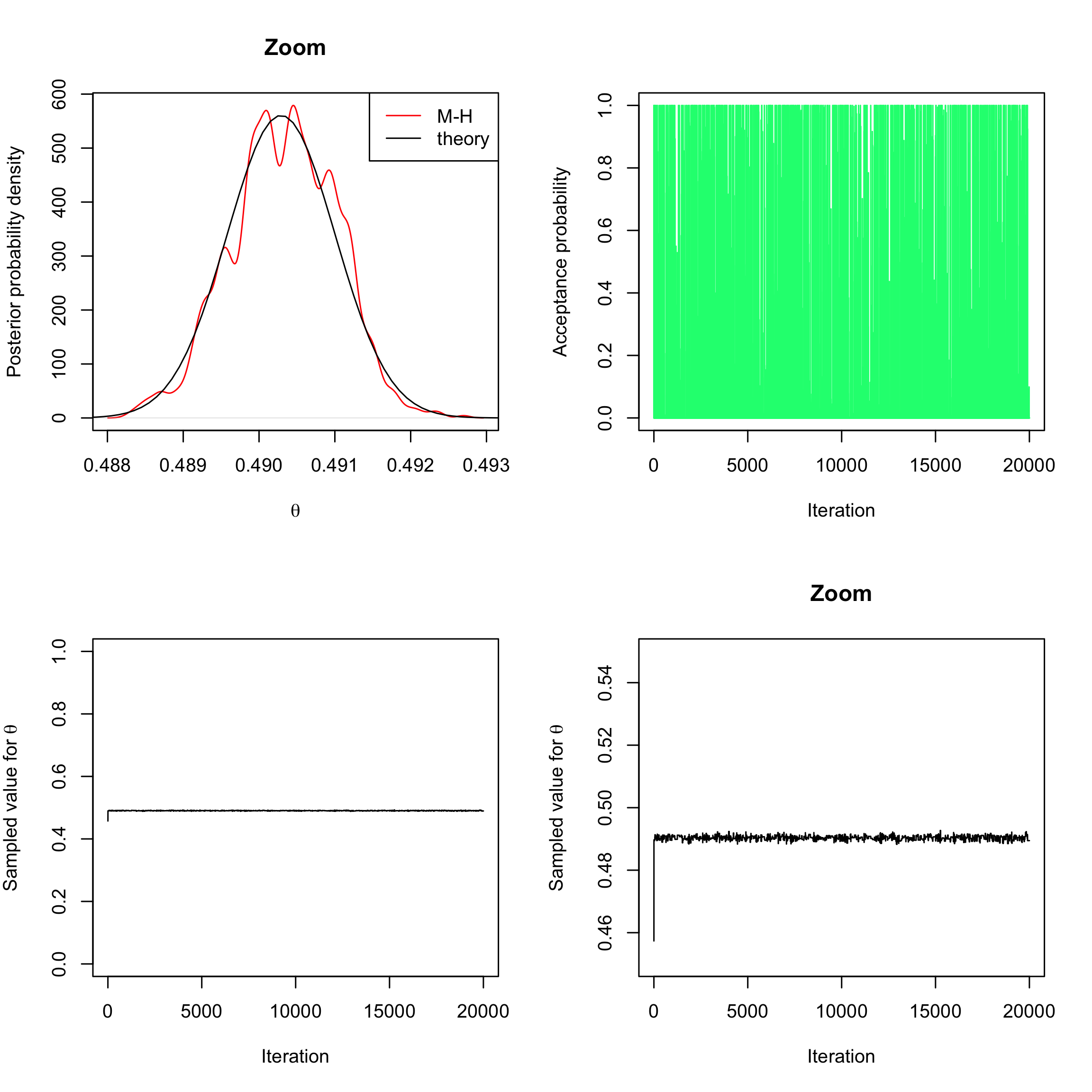
sd=0.02for instance). This means that the proposal is going to change, and is now going to depend on the previous value.
Once again, study the results obtained this way (one can change the width of the random step).post_num_beta_hist <- function(theta, a = 3, b = 3, log = TRUE) { n <- 493472 #number of trials (births) S <- 241945 #number of success (feminine births) if (log) { num <- (a + S - 1) * log(theta) + (b + n - S - 1) * log(1 - theta) } else { num <- theta^(a + S - 1) * (1 - theta)^(b + n - S - 1) } return(num) } myMH_betaprior_randomwalk <- function(niter, post_num, a = 3, b = 3) { x_save <- numeric(length = niter) # create a vector of 0s of length niter to store the sampled values alpha <- numeric(length = niter) # create a vector of 0s of length niter to store the acceptance probabilities # initialise x0 x <- runif(n = 1, min = 0, max = 1) # acceptance-rejection loop for (t in 1:niter) { # sample a value from the proposal (random walk) y <- rnorm(1, mean = x, sd = 0.02) # compute acceptance-rejection probability alpha[t] <- min(1, exp(post_num(y, a = a, b = b, log = TRUE) - post_num(x, a = a, b = b, log = TRUE))) # acceptance-rejection step u <- runif(1) if (u <= alpha[t]) { x <- y # accept y and update current value } # save current value x_save[t] <- x } return(list(theta = x_save, alpha = alpha)) } sampleMH <- myMH_betaprior_randomwalk(20000, post_num = post_num_beta_hist) par(mfrow = c(2, 2)) plot(density(sampleMH$theta[-c(1:1000)]), col = "red", ylab = "Posterior probability density", xlab = expression(theta), main = "Zoom") curve(dbeta(x, 241945 + 1, 251527 + 1), from = 0, to = 1, n = 10000, add = TRUE) legend("topright", c("M-H", "theory"), col = c("red", "black"), lty = 1) plot(sampleMH$alpha, type = "h", xlab = "Iteration", ylab = "Acceptance probability", ylim = c(0, 1), col = "springgreen") plot(sampleMH$theta, type = "l", xlab = "Iteration", ylab = expression(paste("Sampled value for ", theta)), ylim = c(0, 1)) plot(sampleMH$theta, type = "l", xlab = "Iteration", main = "Zoom", ylab = expression(paste("Sampled value for ", theta)), ylim = c(0.45, 0.55))