Exercise 8: Bayesian regression
We will now analyze data from the therapeutic clinical trial ALBI-ANRS 070 which compared the efficacy and tolerance of 3 anti-retroviral strategies among HIV-1 positive patients naive of any anti-retroviral treatment 1.
Load the data from the
albianrs_data.csvavailable here (ProTip: setstringsAsFactors = TRUEinread.delim())albi <- read.csv("albianrs_data.csv", stringsAsFactors = TRUE) summary(albi)## PatientID ViralLoad CD4 CD4t CD4sup500 ## ID112 : 7 Min. :1.699 Min. : 149.0 Min. :3.494 No :558 ## ID130 : 7 1st Qu.:2.042 1st Qu.: 377.0 1st Qu.:4.406 Yes:392 ## ID139 : 7 Median :2.712 Median : 465.0 Median :4.644 ## ID15 : 7 Mean :2.907 Mean : 491.8 Mean :4.664 ## ID156 : 7 3rd Qu.:3.692 3rd Qu.: 585.0 3rd Qu.:4.918 ## ID163 : 7 Max. :5.604 Max. :1318.0 Max. :6.025 ## (Other):908 ## Treatment Day Visit ## AZT+3TC:315 Min. : 0.00 Min. :0.000 ## d4t+ddI:322 1st Qu.: 29.00 1st Qu.:1.000 ## switch :313 Median : 84.00 Median :3.000 ## Mean : 86.19 Mean :2.946 ## 3rd Qu.:140.00 3rd Qu.:5.000 ## Max. :256.00 Max. :6.000 ##The following variables are available:
PatientID: Patient IDViralLoad: Plasma viral loadCD4: CD4 T-cell lymphocites rate (in cell/mm3)CD4t: transformed CD4 T-cell lymphocites rate (in cell/mm3) (CD4t=CD41/4)CD4sup500: binary variable indicating wether CD4 cell counts is above 500Treatment: Treatment groupDay: Day of visit (since inclusion)Visit: Visit number
Below is a quantitative summary of the characteristics of those variables.
| Variable | N = 9501 |
|---|---|
| ViralLoad | 2.71 (2.04, 3.69) |
| CD4 | 465 (377, 585) |
| CD4t | 4.64 (4.41, 4.92) |
| CD4sup500 | 392 (41%) |
| Treatment | |
| AZT+3TC | 315 (33%) |
| d4t+ddI | 322 (34%) |
| switch | 313 (33%) |
| Day | 84 (29, 140) |
| Visit | |
| 0 | 146 (15%) |
| 1 | 140 (15%) |
| 2 | 136 (14%) |
| 3 | 130 (14%) |
| 4 | 128 (13%) |
| 5 | 135 (14%) |
| 6 | 135 (14%) |
| 1 Median (IQR) or n (%) | |
NB: for simplicity, NA values have been omitted.
First, we want to explain the CD4 rate (as a transformed variable) from the viral load
Display
CD4tin scatterplot as a function of theViralLoadand color the points according toTreatment.library(ggplot2) library(MetBrewer) ggplot(albi, aes(y = CD4t, x = ViralLoad, color = Treatment)) + geom_point(alpha = 0.7) + MetBrewer::scale_color_met_d("Java") + theme_bw()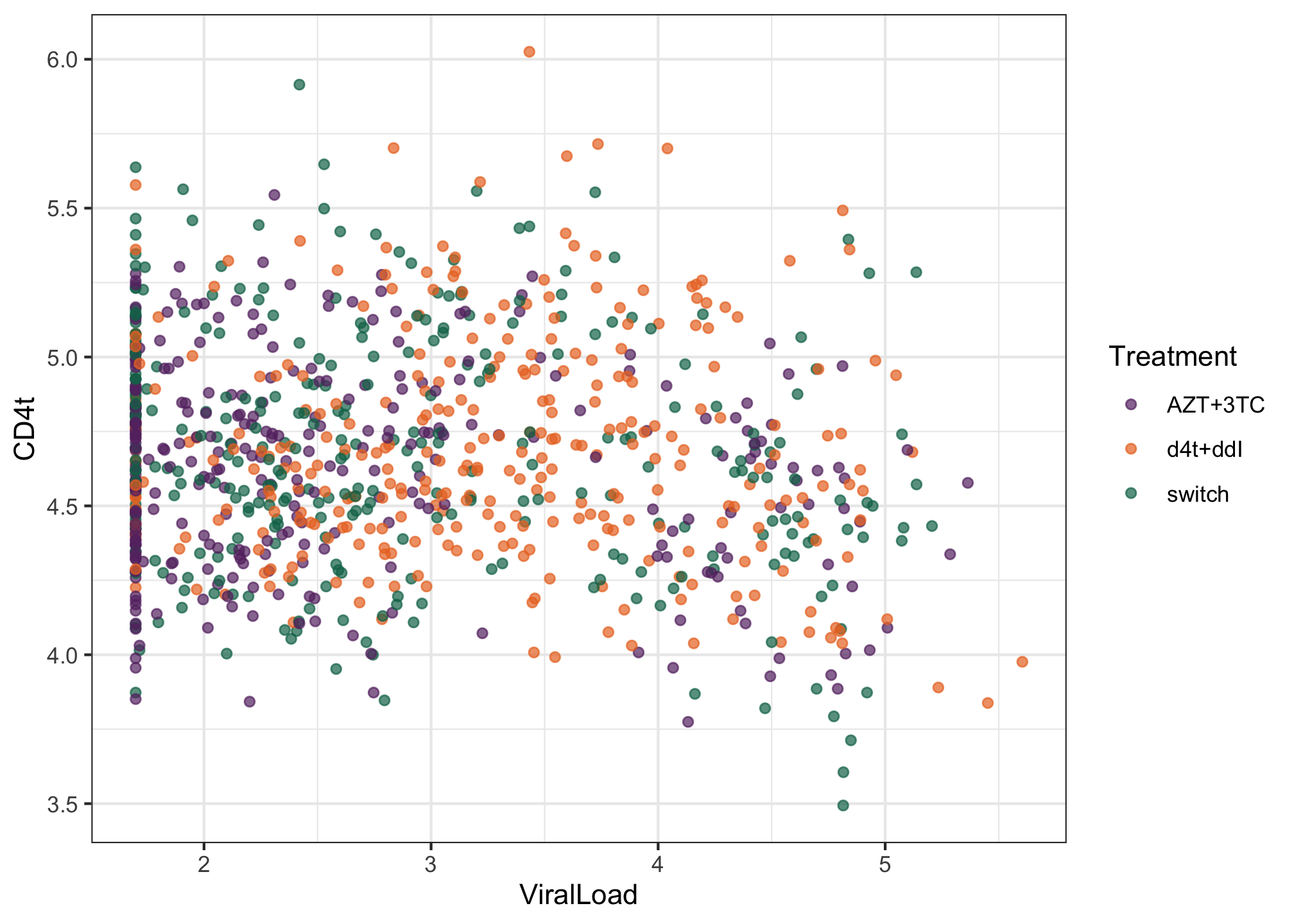
Write down the Bayesian mathematical model corresponding to a linear regression of
CD4tagainst theViralLoad.
I) Question of interest:
How does the viral load linearly explains the (transformed) CD4 T-cell rate ?
II) Sampling model:
LetWrite the corresponding
BUGSmodel and save it in an external.txtfile.# Fixed effect linear regression Albi-ANRS model{ # Likelihood for (i in 1:N){ CD4t[i]~dnorm(mu[i], tau) mu[i] <- beta0 + beta1*VL[i] } # Priors beta0~dnorm(0,0.0001) beta1~dnorm(0,0.0001) # proper but very flat (vague: weakly informative) tau~dgamma(0.0001,0.0001) # proper but very flat (vague: weakly informative) # Parameters of interest sigma <- pow(tau, -0.5) }Create the corresponding
jagsobject in and generate a Monte Carlo sample of size 1000 for the 3 parameters of interestlibrary(rjags) albi_fixed_jags <- jags.model("albiBUGSmodel_fixed.txt", data = list(CD4t = albi$CD4t, N = nrow(albi), VL = albi$ViralLoad), n.chains = 3)## Compiling model graph ## Resolving undeclared variables ## Allocating nodes ## Graph information: ## Observed stochastic nodes: 950 ## Unobserved stochastic nodes: 3 ## Total graph size: 3195 ## ## Initializing modelres_albi_fixed <- coda.samples(model = albi_fixed_jags, variable.names = c("beta0", "beta1", "sigma"), n.iter = 1000)Before interpreting the results, check the convergence. What do you notice ?
library(coda) plot(res_albi_fixed)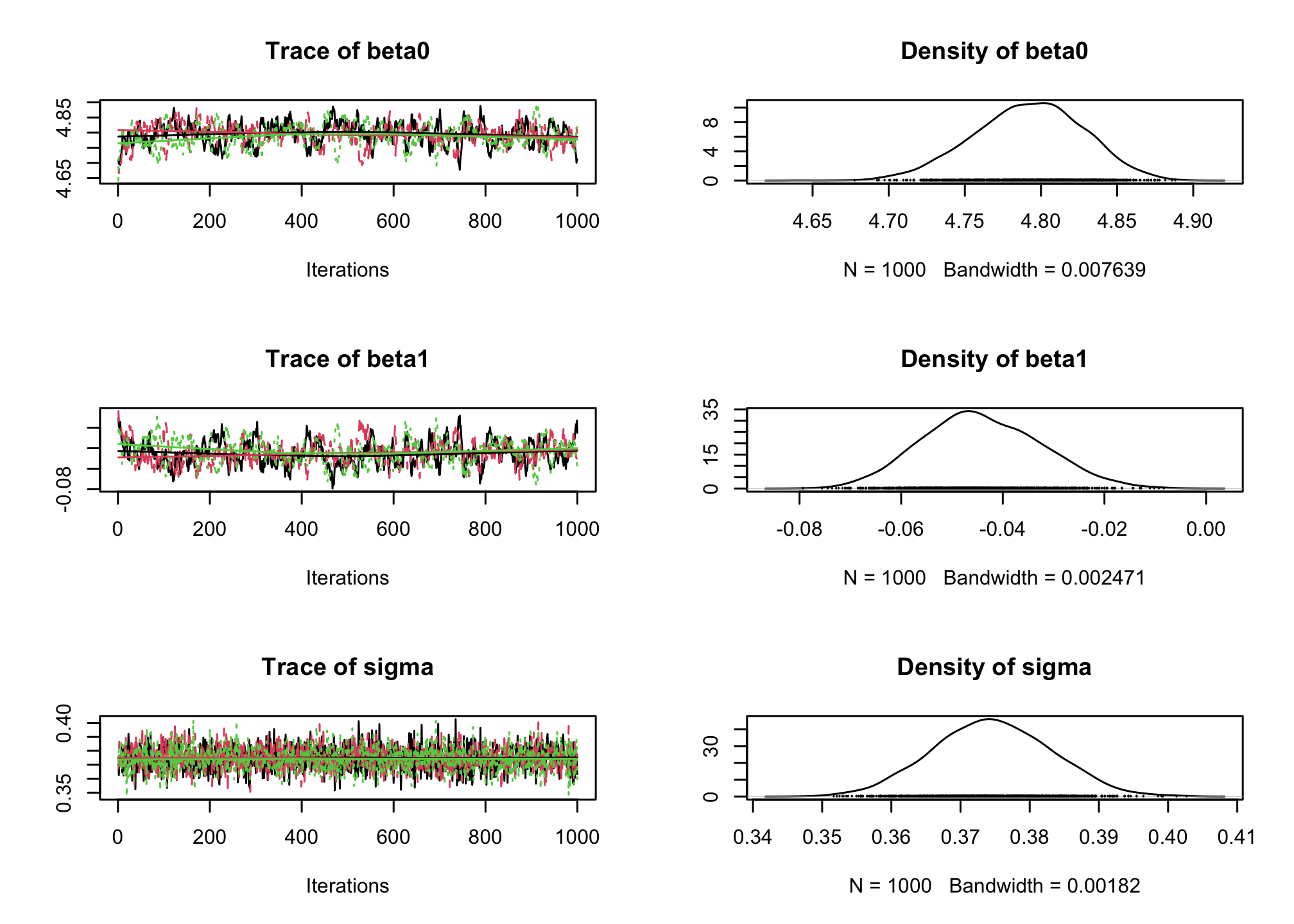
gelman.plot(res_albi_fixed)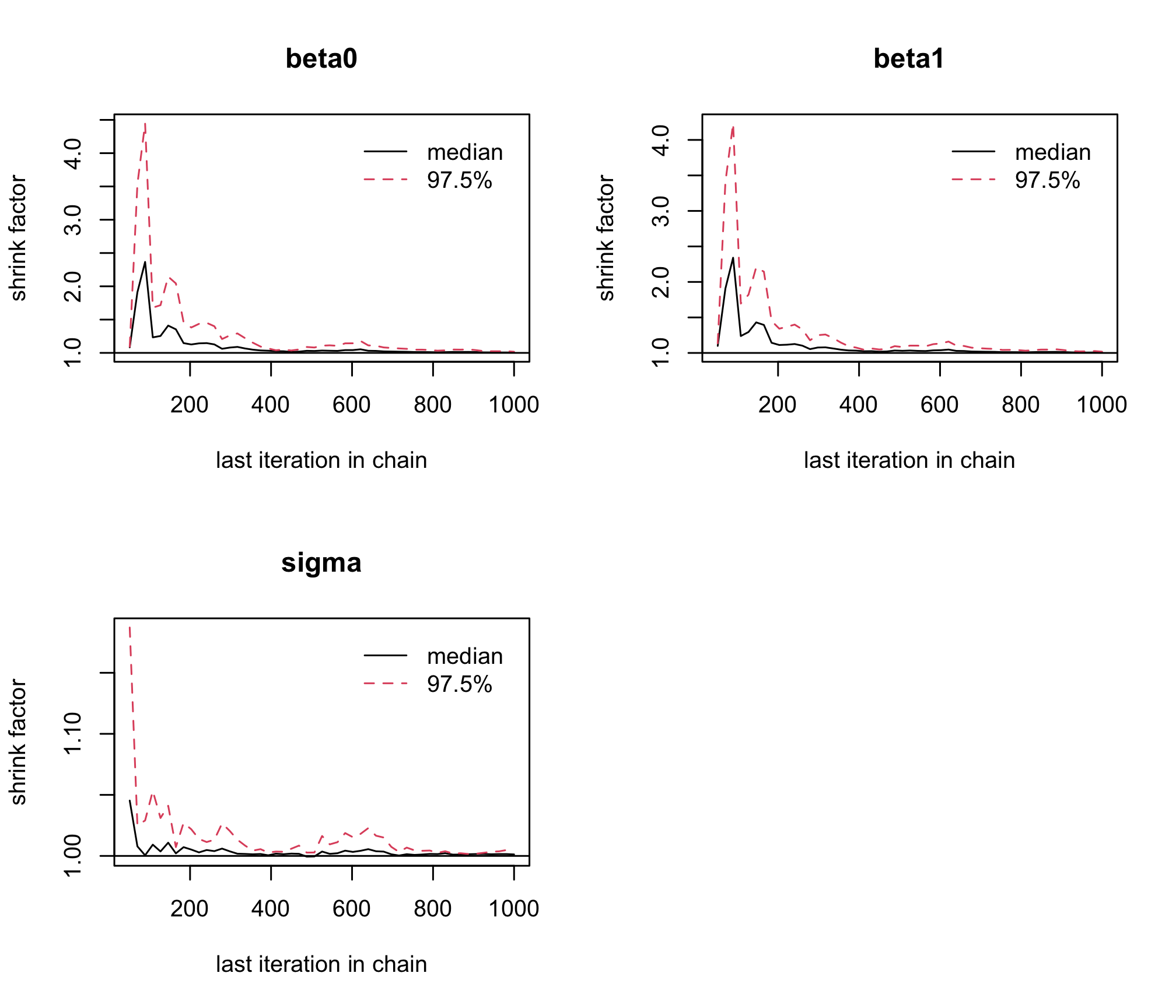
acfplot(res_albi_fixed)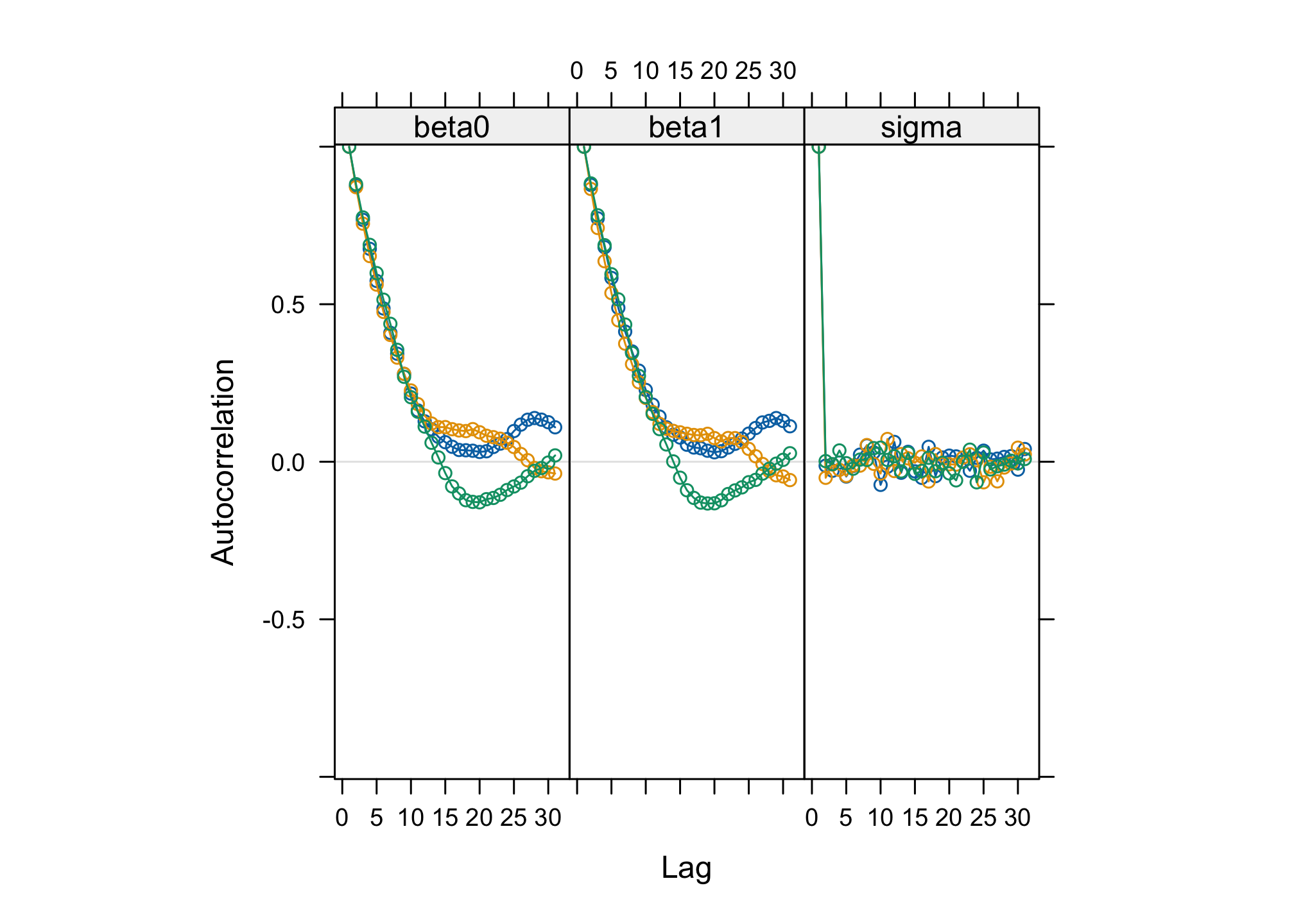
par(mfrow = c(3, 3)) cumuplot(res_albi_fixed, ask = FALSE, auto.layout = FALSE)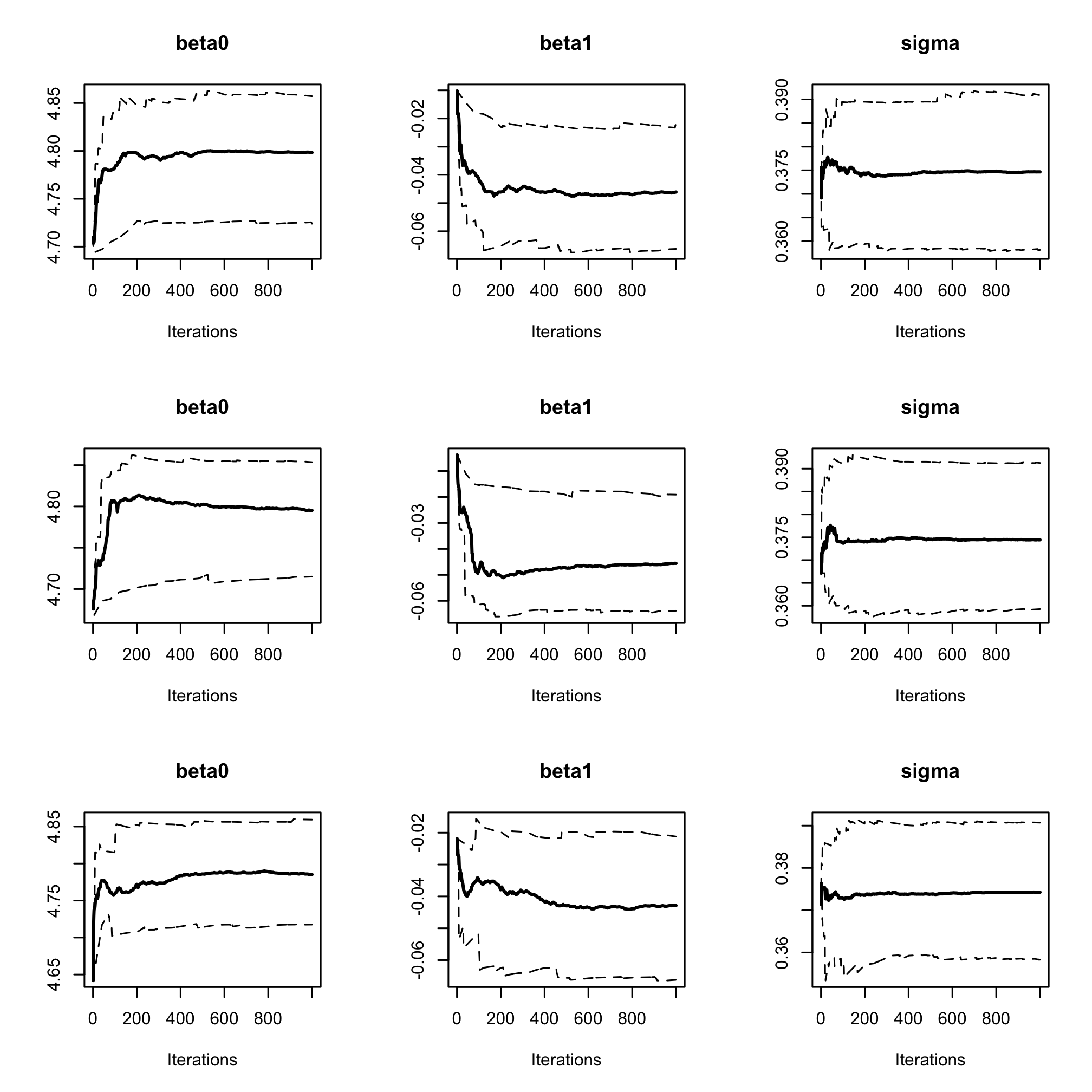
Using the
windowfunction, remove the 500 first iterations as burn-in to reach Markov Chain convergence to its stationary distribution (the posterior). Is it sufficient to solve convergence issues ?res_albi_fixed2 <- window(res_albi_fixed, start = 501) gelman.plot(res_albi_fixed2)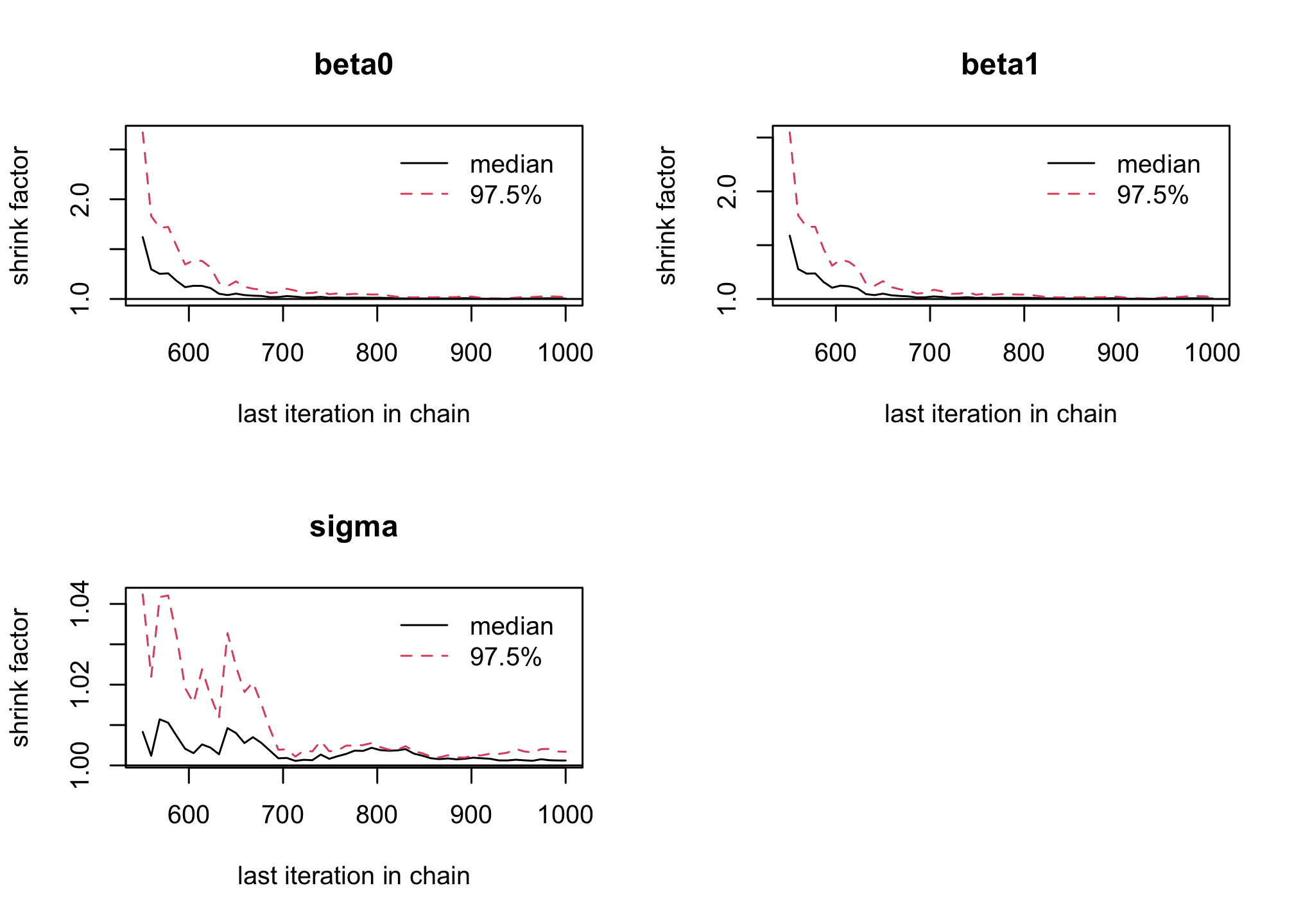
acfplot(res_albi_fixed2)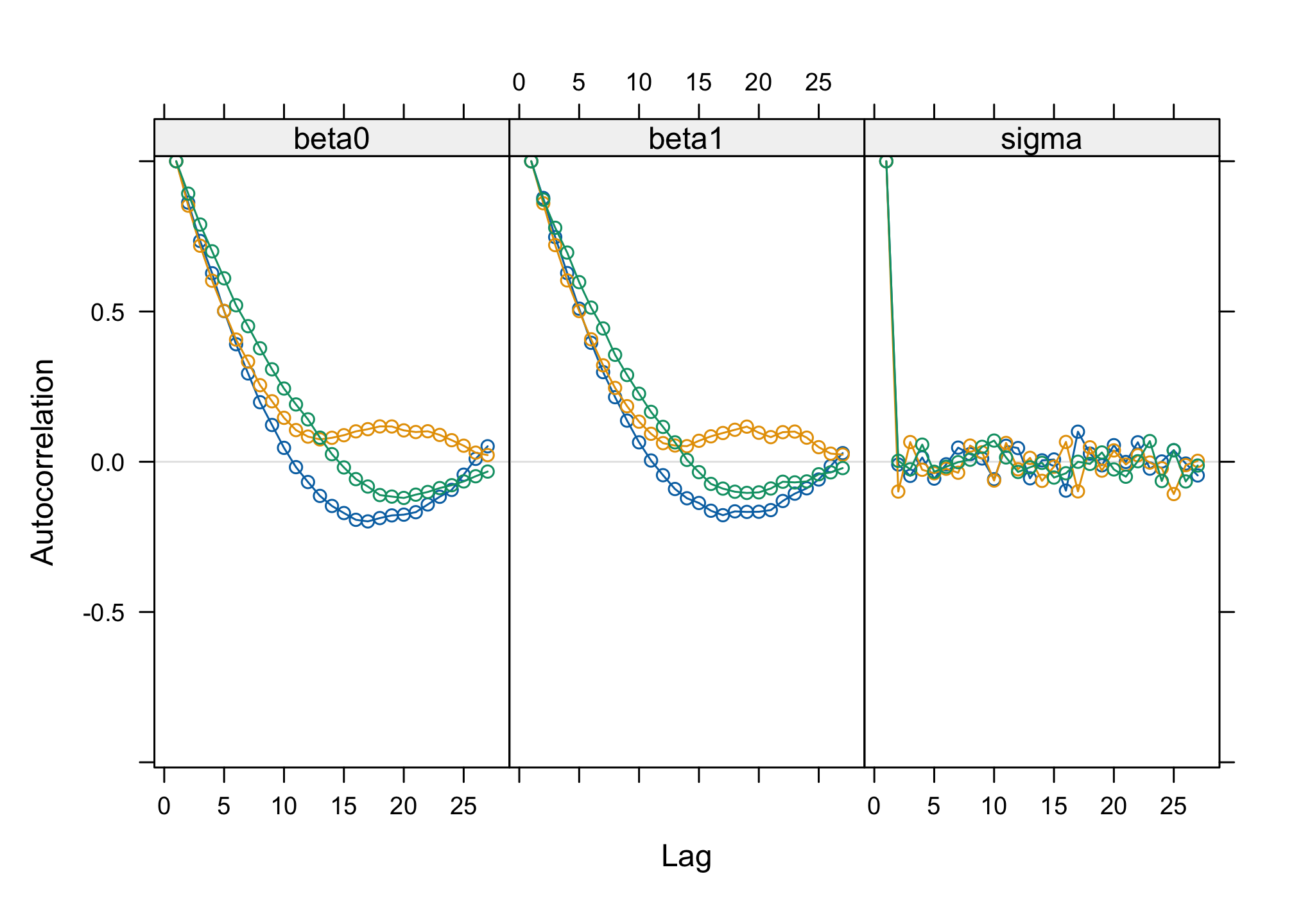
Check the effective sample size of the Monte Carlo sample with the
effectiveSize()function. Reduce auto-correlation by increasing thethinparameter to 10 incoda.samples. Check the impact on the effective sample`?`(effectiveSize) effectiveSize(res_albi_fixed2)## beta0 beta1 sigma ## 119.0255 117.9688 1608.7182albi_fixed_jags <- jags.model("albiBUGSmodel_fixed.txt", data = list(CD4t = albi$CD4t, N = nrow(albi), VL = albi$ViralLoad), n.chains = 3)## Compiling model graph ## Resolving undeclared variables ## Allocating nodes ## Graph information: ## Observed stochastic nodes: 950 ## Unobserved stochastic nodes: 3 ## Total graph size: 3195 ## ## Initializing modelres_albi_fixed_thin <- coda.samples(model = albi_fixed_jags, variable.names = c("beta0", "beta1", "sigma"), n.iter = 10000, thin = 10) res_albi_fixed_thin <- window(res_albi_fixed_thin, start = 5001) effectiveSize(res_albi_fixed_thin)## beta0 beta1 sigma ## 697.3918 645.3890 1567.0829plot(res_albi_fixed_thin)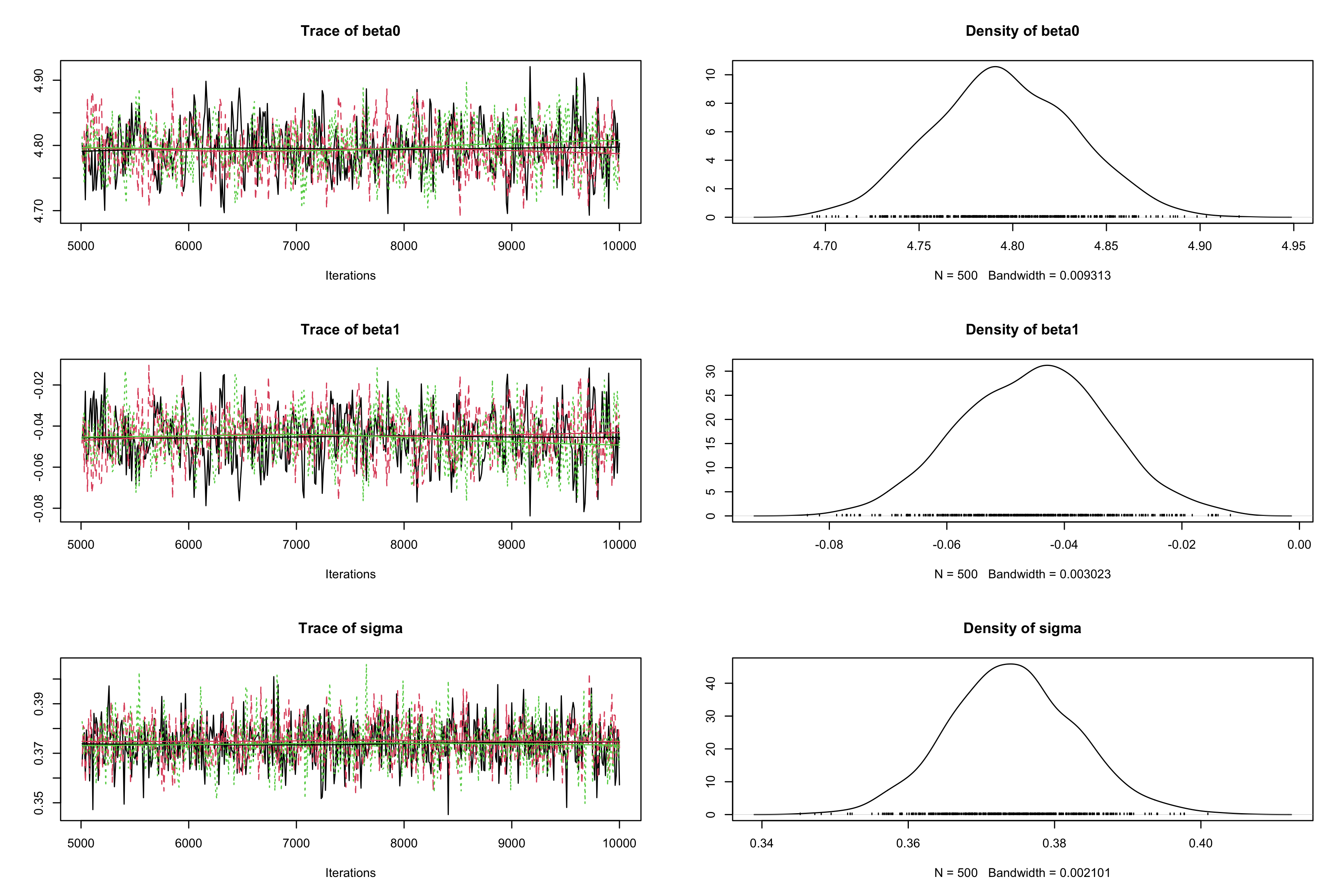
gelman.plot(res_albi_fixed_thin)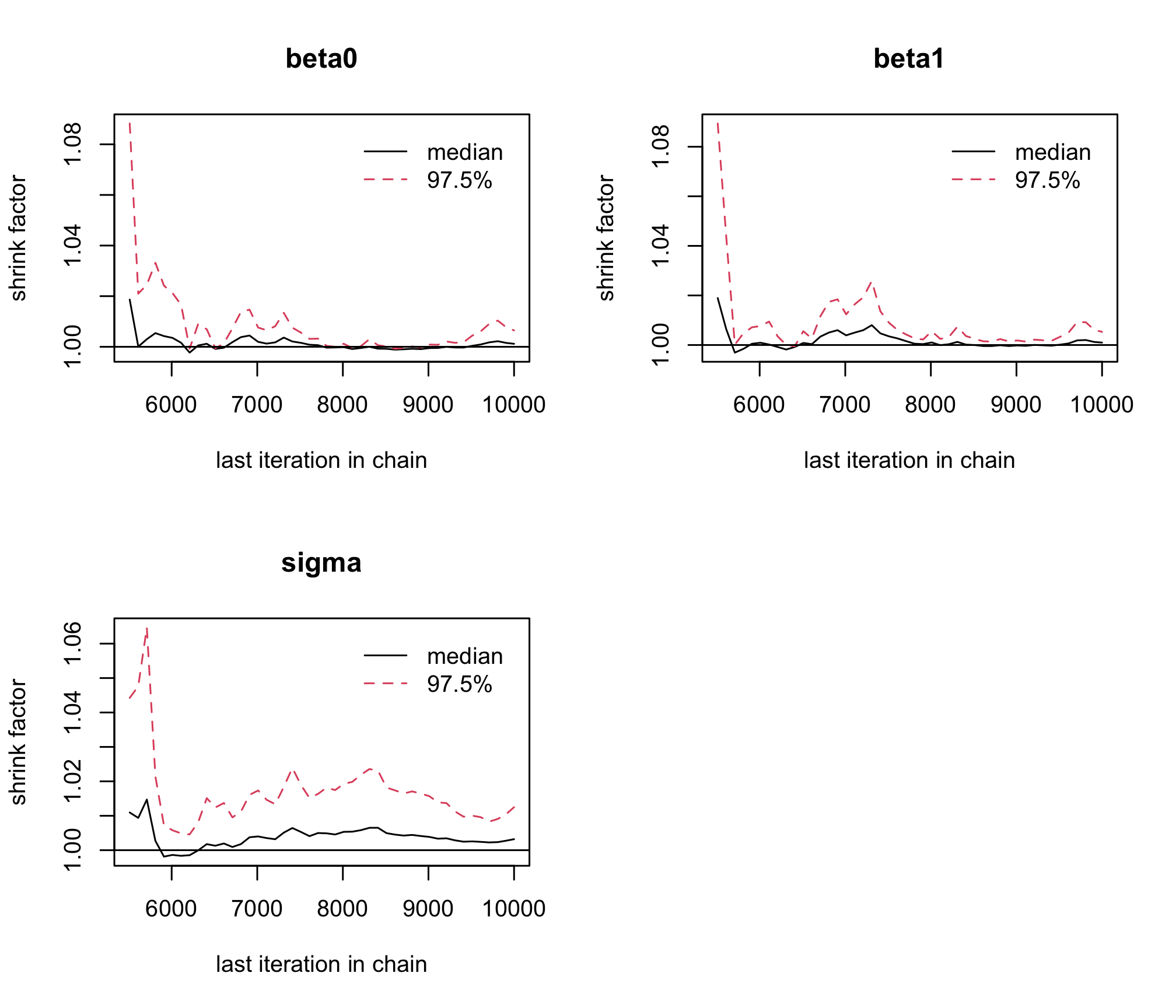
acfplot(res_albi_fixed_thin)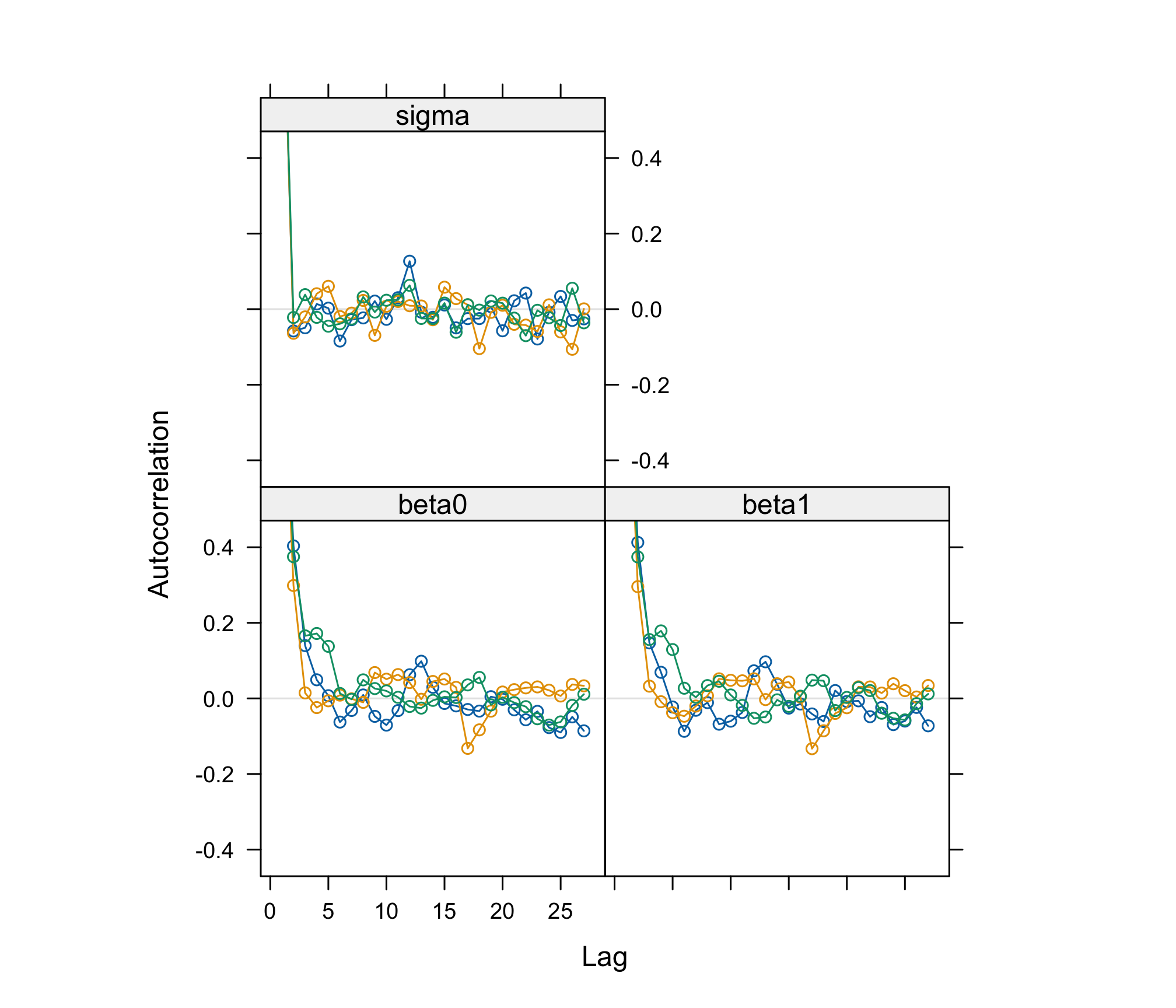
par(mfrow = c(3, 3)) cumuplot(res_albi_fixed_thin, ask = FALSE, auto.layout = FALSE)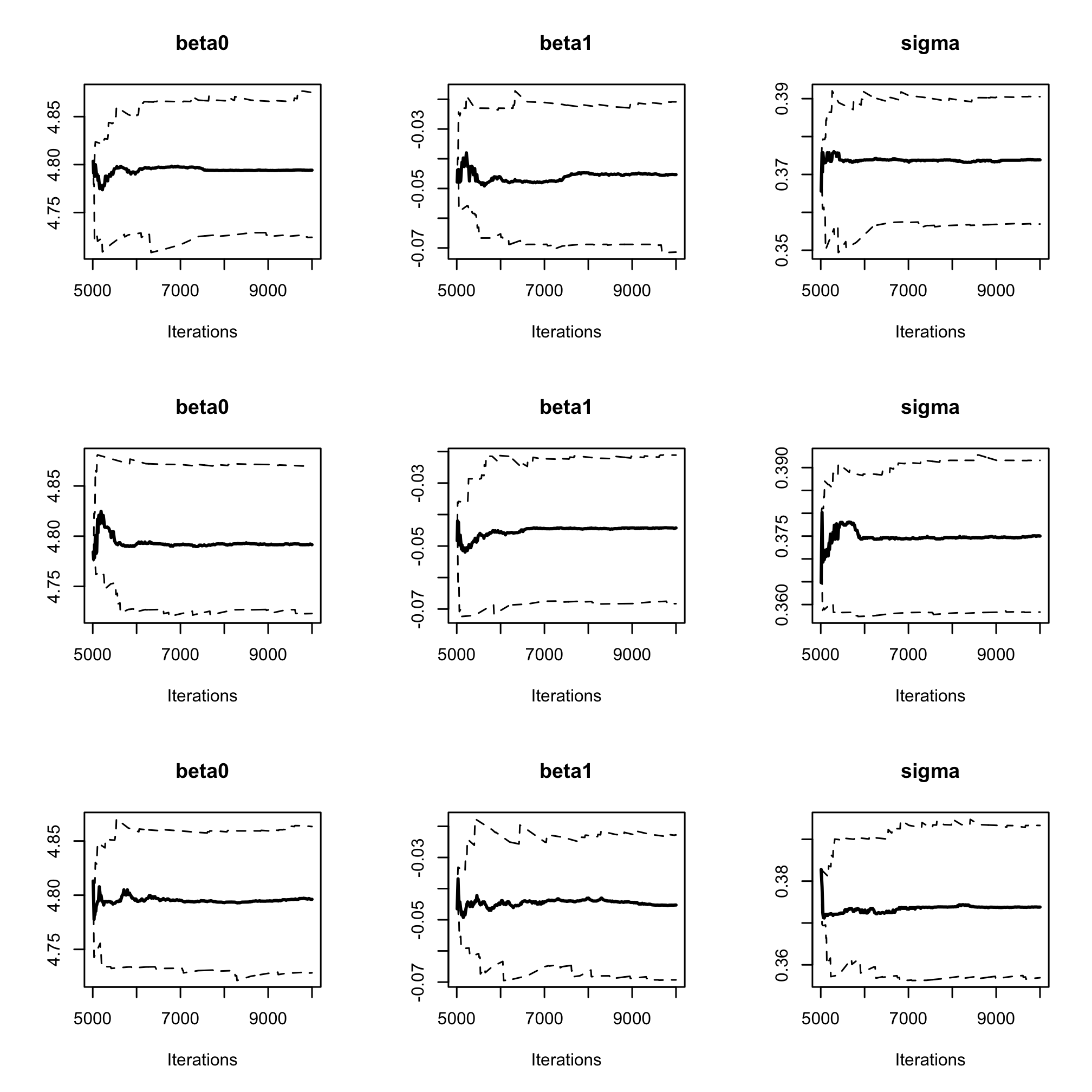
summary(res_albi_fixed_thin)## ## Iterations = 5010:10000 ## Thinning interval = 10 ## Number of chains = 3 ## Sample size per chain = 500 ## ## 1. Empirical mean and standard deviation for each variable, ## plus standard error of the mean: ## ## Mean SD Naive SE Time-series SE ## beta0 4.79520 0.037932 0.0009794 0.0014860 ## beta1 -0.04529 0.012314 0.0003179 0.0004939 ## sigma 0.37429 0.008766 0.0002263 0.0002219 ## ## 2. Quantiles for each variable: ## ## 2.5% 25% 50% 75% 97.5% ## beta0 4.72417 4.76969 4.79415 4.82216 4.86880 ## beta1 -0.06898 -0.05388 -0.04489 -0.03683 -0.02111 ## sigma 0.35744 0.36842 0.37413 0.37989 0.39242
Compare those results with a frequentist analysis.
freq_mod <- lm(CD4t ~ ViralLoad, data = albi) summary(freq_mod)## ## Call: ## lm(formula = CD4t ~ ViralLoad, data = albi) ## ## Residuals: ## Min 1Q Median 3Q Max ## -1.0828 -0.2604 -0.0197 0.2449 1.3856 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 4.79635 0.03696 129.8 < 2e-16 *** ## ViralLoad -0.04564 0.01201 -3.8 0.000154 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 0.3742 on 948 degrees of freedom ## Multiple R-squared: 0.01501, Adjusted R-squared: 0.01397 ## F-statistic: 14.44 on 1 and 948 DF, p-value: 0.0001537confint(freq_mod)## 2.5 % 97.5 % ## (Intercept) 4.72381166 4.86888881 ## ViralLoad -0.06921147 -0.02207249Perform a Bayesian logistic regression to study the impact of the treatment on the binary outcome
CD4sup500adjusted on the viral load. Once you have a first estimation, try adding an interaction between the treatment and the viral load.# Fixed effect logistic regression Albi-ANRS model{ # likelihood for (i in 1:N){ CD4sup500[i] ~ dbern(proba[i]) logit(proba[i]) <- beta0 + beta1*VL[i] + beta2*Tt_switch[i] + beta3*d4t[i] + beta4*VL[i]*d4t[i] + beta5*VL[i]*Tt_switch[i] } #Priors beta0~dnorm(0,0.0001) beta1~dnorm(0,0.0001) beta2~dnorm(0,0.0001) beta3~dnorm(0,0.0001) beta4~dnorm(0,0.0001) beta5~dnorm(0,0.0001) #ORs OR_VL <- exp(beta1) OR_switch <- exp(beta2) OR_d4t <- exp(beta3) OR_d4tVL <- exp(beta4) OR_swVL <- exp(beta5) }library(rjags) albi_logis_jags <- jags.model("albiBUGSmodel_logistic_fixed.txt", data = list(CD4sup500 = 1 * (albi$CD4sup500 == "Yes"), N = nrow(albi), VL = albi$ViralLoad, d4t = 1 * (albi$Treatment == "d4t+ddI"), Tt_switch = 1 * (albi$Treatment == "switch")), n.chains = 3)## Compiling model graph ## Resolving undeclared variables ## Allocating nodes ## Graph information: ## Observed stochastic nodes: 950 ## Unobserved stochastic nodes: 6 ## Total graph size: 7299 ## ## Initializing modelres_albi_logis <- coda.samples(model = albi_logis_jags, variable.names = c("OR_VL", "OR_switch", "OR_d4t", "OR_d4tVL", "OR_swVL"), n.iter = 10000) res_albi_logis_burnt <- window(res_albi_logis, start = 5001) plot(res_albi_logis_burnt)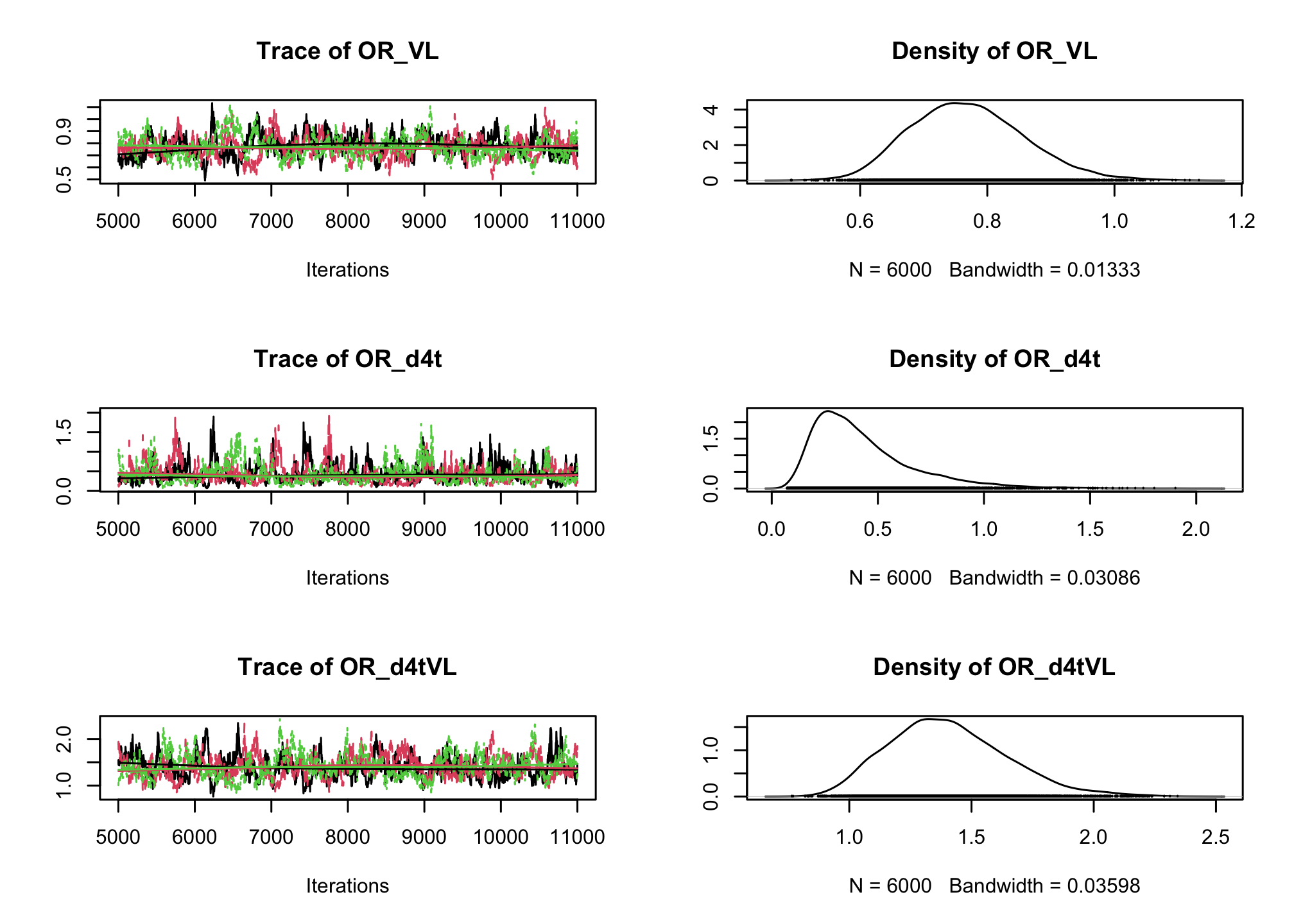
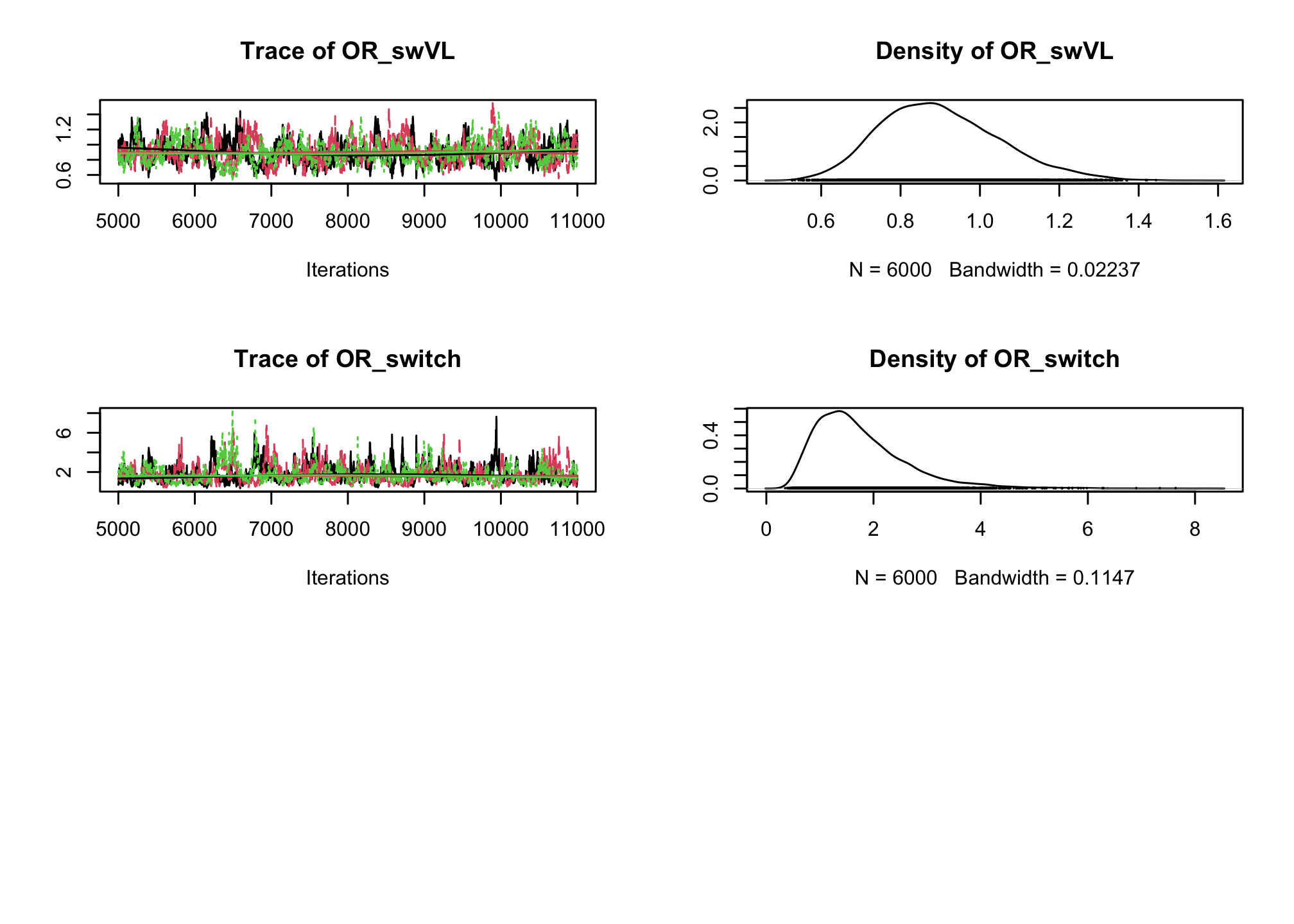
summary(res_albi_logis_burnt)## ## Iterations = 5001:11000 ## Thinning interval = 1 ## Number of chains = 3 ## Sample size per chain = 6000 ## ## 1. Empirical mean and standard deviation for each variable, ## plus standard error of the mean: ## ## Mean SD Naive SE Time-series SE ## OR_VL 0.7700 0.08925 0.0006653 0.006531 ## OR_d4t 0.4285 0.23862 0.0017785 0.016362 ## OR_d4tVL 1.4041 0.24089 0.0017955 0.016002 ## OR_swVL 0.9012 0.14977 0.0011163 0.008414 ## OR_switch 1.7516 0.86748 0.0064659 0.048294 ## ## 2. Quantiles for each variable: ## ## 2.5% 25% 50% 75% 97.5% ## OR_VL 0.6134 0.7067 0.7654 0.8275 0.9595 ## OR_d4t 0.1362 0.2576 0.3688 0.5344 1.0461 ## OR_d4tVL 0.9967 1.2323 1.3850 1.5552 1.9364 ## OR_swVL 0.6442 0.7924 0.8876 0.9970 1.2291 ## OR_switch 0.6214 1.1350 1.5697 2.1640 3.9656summary(glm(CD4sup500 == "Yes" ~ ViralLoad * Treatment, family = "binomial", data = albi))## ## Call: ## glm(formula = CD4sup500 == "Yes" ~ ViralLoad * Treatment, family = "binomial", ## data = albi) ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.3279 0.3241 1.012 0.3117 ## ViralLoad -0.2556 0.1197 -2.135 0.0328 * ## Treatmentd4t+ddI -0.9409 0.5338 -1.763 0.0779 . ## Treatmentswitch 0.4836 0.4807 1.006 0.3143 ## ViralLoad:Treatmentd4t+ddI 0.3084 0.1737 1.776 0.0757 . ## ViralLoad:Treatmentswitch -0.1303 0.1690 -0.771 0.4409 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for binomial family taken to be 1) ## ## Null deviance: 1287.8 on 949 degrees of freedom ## Residual deviance: 1270.9 on 944 degrees of freedom ## AIC: 1282.9 ## ## Number of Fisher Scoring iterations: 4So far we have ignored the longitudinal nature of our measurements. Now, lets add a random intercept in our logistic regression to account for the intra-patient correlation across visits.
ggplot(albi, aes(y = CD4, x = Day, color = Treatment)) + geom_hline(aes(yintercept = 500), color = "grey35", linetype = 2) + geom_line(aes(group = PatientID), alpha = 0.3) + MetBrewer::scale_color_met_d("Java") + theme_bw()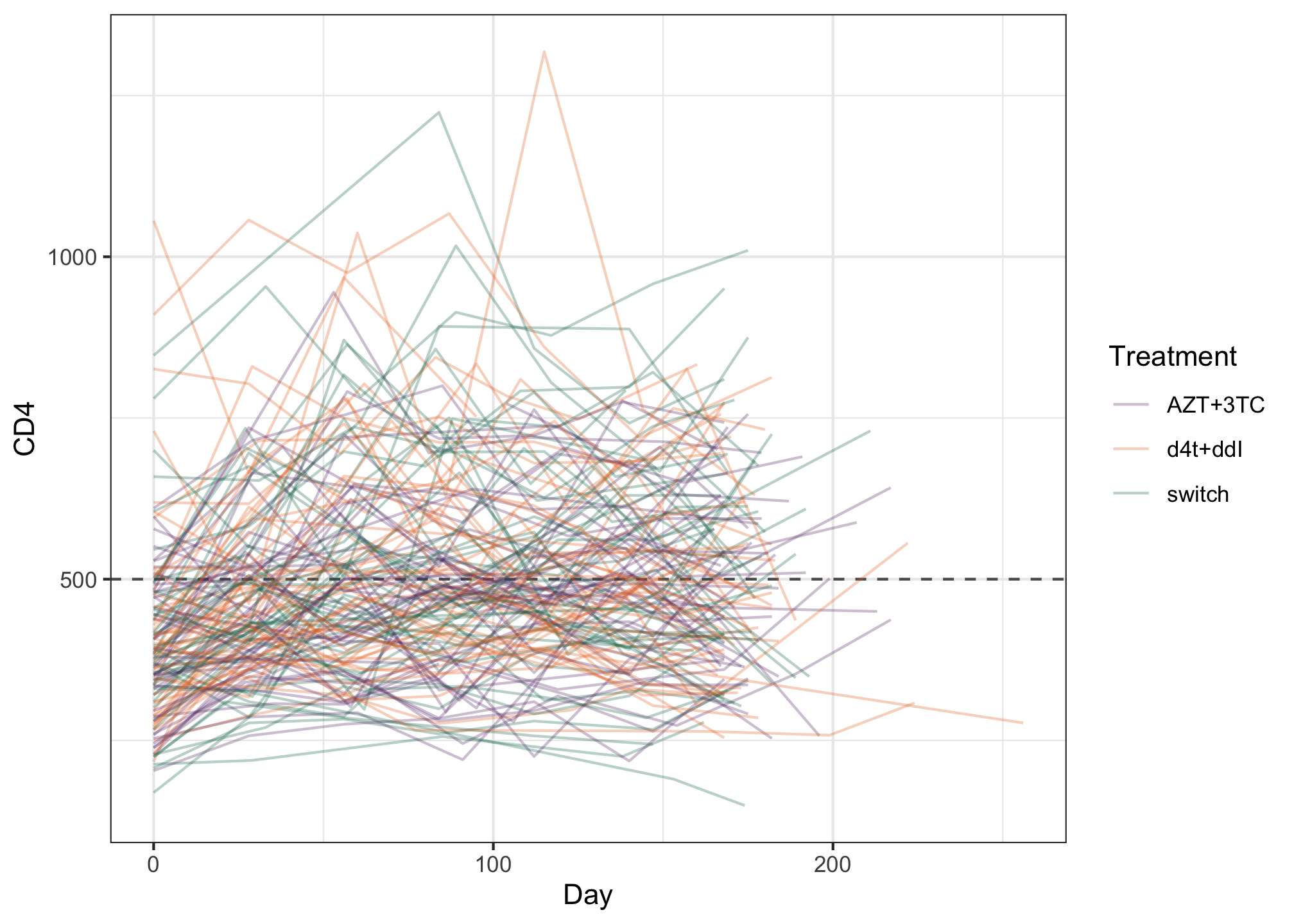
# Mixed effect logistic regression Albi-ANRS model{ # likelihood for (i in 1:N){ CD4sup500[i] ~ dbern(proba[i]) logit(proba[i]) <- beta0 + b0[PAT_ID[i]] + beta1*VL[i] + beta2*Tt_switch[i] + beta3*d4t[i] + beta4*VL[i]*d4t[i] + beta5*VL[i]*Tt_switch[i] } for (j in 1:Npat){ b0[j]~dnorm(0, tau_b0) } #Priors beta0~dnorm(0,0.0001) beta1~dnorm(0,0.0001) beta2~dnorm(0,0.0001) beta3~dnorm(0,0.0001) beta4~dnorm(0,0.0001) beta5~dnorm(0,0.0001) tau_b0~dgamma(0.001,0.001) #ORs OR_VL <- exp(beta1) OR_switch <- exp(beta2) OR_d4t <- exp(beta3) OR_d4tVL <- exp(beta4) OR_swVL <- exp(beta5) sigma_RI <- pow(tau_b0, -0.5) }library(rjags) albi_logis_RI_jags <- jags.model("albiBUGSmodel_logistic_RandomInt.txt", data = list(CD4sup500 = 1 * (albi$CD4sup500 == "Yes"), N = nrow(albi), VL = albi$ViralLoad, d4t = 1 * (albi$Treatment == "d4t+ddI"), Tt_switch = 1 * (albi$Treatment == "switch"), Npat = length(levels(albi$PatientID)), PAT_ID = as.numeric(albi$PatientID)), n.chains = 3)## Compiling model graph ## Resolving undeclared variables ## Allocating nodes ## Graph information: ## Observed stochastic nodes: 950 ## Unobserved stochastic nodes: 156 ## Total graph size: 8670 ## ## Initializing modelres_albi_logis_RI <- coda.samples(model = albi_logis_RI_jags, variable.names = c("OR_VL", "OR_switch", "OR_d4t", "OR_d4tVL", "OR_swVL", "sigma_RI"), n.iter = 10000) res_albi_logis_RI_burnt <- window(res_albi_logis_RI, start = 1001) plot(res_albi_logis_RI_burnt)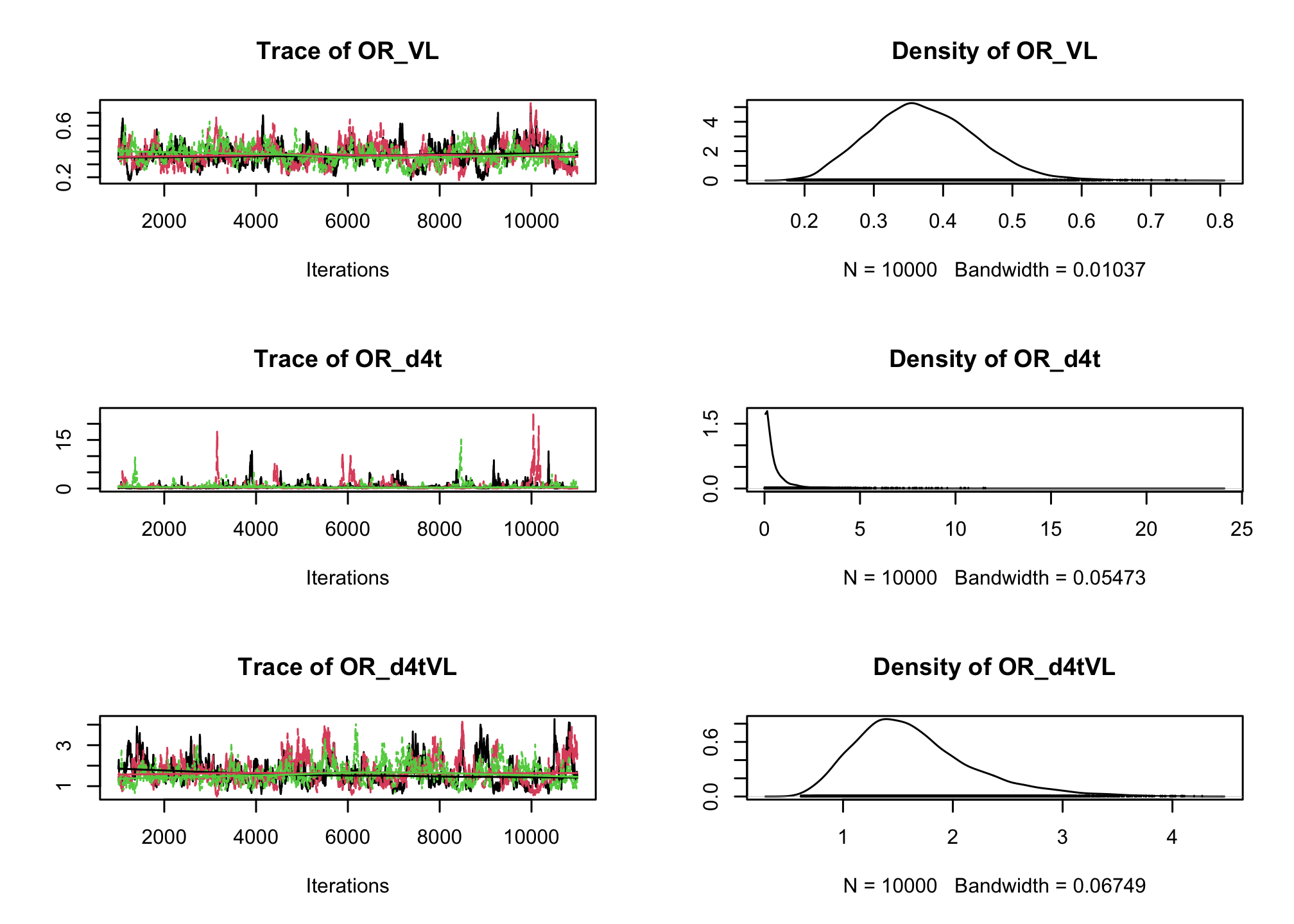
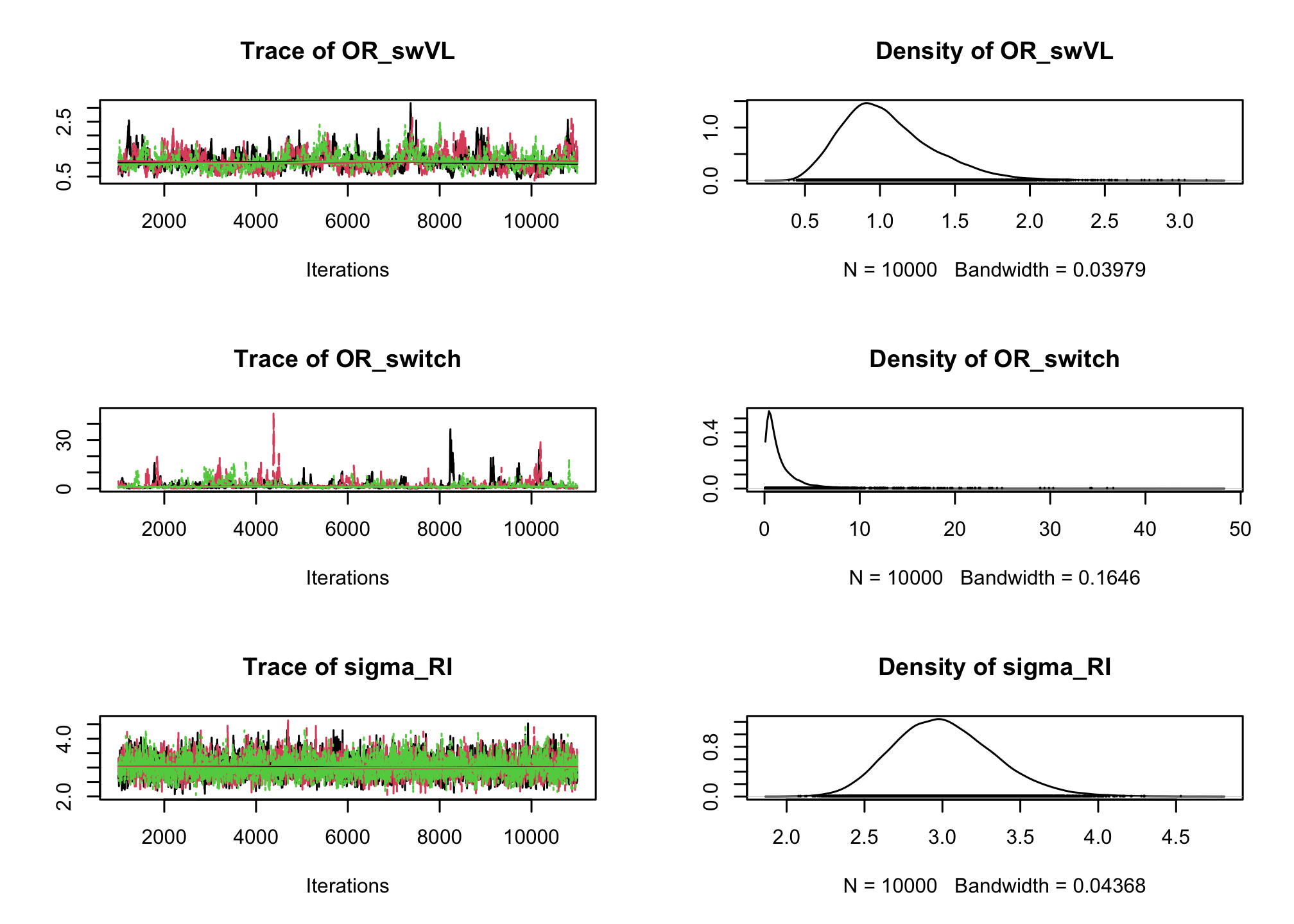
summary(res_albi_logis_RI_burnt)## ## Iterations = 1001:11000 ## Thinning interval = 1 ## Number of chains = 3 ## Sample size per chain = 10000 ## ## 1. Empirical mean and standard deviation for each variable, ## plus standard error of the mean: ## ## Mean SD Naive SE Time-series SE ## OR_VL 0.3725 0.07691 0.000444 0.004670 ## OR_d4t 0.6352 1.13532 0.006555 0.061109 ## OR_d4tVL 1.6485 0.54363 0.003139 0.034870 ## OR_swVL 1.0544 0.31312 0.001808 0.017495 ## OR_switch 1.8321 2.41479 0.013942 0.122489 ## sigma_RI 3.0162 0.32610 0.001883 0.006224 ## ## 2. Quantiles for each variable: ## ## 2.5% 25% 50% 75% 97.5% ## OR_VL 0.2366 0.3183 0.3677 0.4221 0.5325 ## OR_d4t 0.0321 0.1370 0.3070 0.6808 3.3165 ## OR_d4tVL 0.8316 1.2657 1.5620 1.9363 2.9791 ## OR_swVL 0.5749 0.8327 1.0062 1.2281 1.7807 ## OR_switch 0.1492 0.5436 1.0792 2.1791 8.1893 ## sigma_RI 2.4420 2.7881 2.9946 3.2221 3.7113library(lme4) summary(lme4::glmer(CD4sup500 == "Yes" ~ (1 | PatientID) + ViralLoad * Treatment, family = "binomial", data = albi))## Generalized linear mixed model fit by maximum likelihood (Laplace ## Approximation) [glmerMod] ## Family: binomial ( logit ) ## Formula: CD4sup500 == "Yes" ~ (1 | PatientID) + ViralLoad * Treatment ## Data: albi ## ## AIC BIC logLik -2*log(L) df.resid ## 971.8 1005.8 -478.9 957.8 943 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -2.9273 -0.4021 -0.1947 0.4119 3.9688 ## ## Random effects: ## Groups Name Variance Std.Dev. ## PatientID (Intercept) 7.317 2.705 ## Number of obs: 950, groups: PatientID, 149 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.901075 0.689952 2.755 0.00586 ** ## ViralLoad -0.955590 0.212725 -4.492 7.05e-06 *** ## Treatmentd4t+ddI -1.177377 1.103323 -1.067 0.28592 ## Treatmentswitch 0.101717 1.000232 0.102 0.91900 ## ViralLoad:Treatmentd4t+ddI 0.439794 0.308829 1.424 0.15443 ## ViralLoad:Treatmentswitch 0.008679 0.291172 0.030 0.97622 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) VirlLd Trt4+I Trtmnt VL:T4+ ## ViralLoad -0.780 ## Trtmntd4t+I -0.616 0.473 ## Trtmntswtch -0.679 0.521 0.423 ## VrlLd:Tr4+I 0.518 -0.657 -0.821 -0.355 ## VrlLd:Trtmn 0.549 -0.695 -0.341 -0.786 0.475
Jean-Michel Molina et al. “The ALBI Trial: A Randomized Controlled Trial Comparing Stavudine Plus Didanosine with Zidovudine Plus Lamivudine and a Regimen Alternating Both Combinations in Previously Untreated Patients Infected with Human Immunodeficiency Virus,” The Journal of Infectious Diseases 180, no. 2 (1999): 351–358, doi:10.1086/314891.↩︎